
Gloria Culver
she/her/hers
Professor
PhD
- Office Location
- 484 Hutchison
- Telephone
- (585) 274-2567
Research Overview
Assembly of the E. coli 30S ribosomal subunit.
Ribonucleoprotein particles (RNPs) play essential roles in virtually all aspects of cell growth and regulation, ranging from protein synthesis and transport to RNA processing and DNA replication. Understanding the dynamic assembly and function of these particles is central to understanding the molecular mechanisms that govern these fundamental cellular processes.
The direct correlation between the rates of E. coli growth and ribosome biogenesis indicates that an accurately assembled and functional ribosome is one of the most important cellular organelles. Ribosomes are amazingly complex ribonucleoprotein machines, capable of interacting with a multitude of substrates and factors and accurately converting genotype to phenotype.
Recent advances in understanding the structure of ribosomes and ribosomal subunits in various states has greatly increased our appreciation of these massive macromolecular machines. Given these structures as an end-point, our lab is interested in understanding how the small (30S) subunit of E. coli ribosomes is assembled from the large 16S ribosomal RNA (rRNA) and and twenty-one individual proteins (Figure 1).
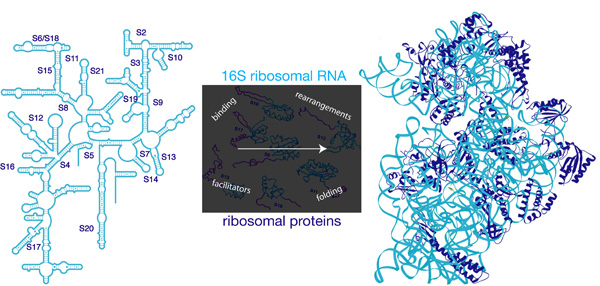
A variety of biochemical, structural and genetic approaches will be directed at understanding the pathway, dynamics and components involved in 30S subunit assembly; this process must ultimately result in a functional 30S subunit that plays an integral role in maintenance of fidelity during translation of the mRNA code into functional protein products.
The genes encoding all 21 small subunit ribosomal proteins (S1-S21) from E. coli have been cloned, and the proteins have been overexpressed and individually purified. Conditions for in vitro reconstitution of functional 30S subunits from a complete set of recombinant proteins and 16S rRNA have been determined. These tools allow detailed analysis of 30S subunit assembly and small subunit ribosomal protein function. The following list is a brief outline of some of the areas our group is pursuing.
1) Probing 16S rRNA Folding during Ribosome Assembly
Mapping the folding events that occur during assembly of ribonucleoprotein particles may allow an understanding of how functional sites are formed in the full complex. Protein-induced RNA conformation changes can be monitored during the course of assembly by using the recombinant proteins as site-directed probes. Changes in the folding of the RNA backbone at different stages of assembly will be identified. By building different assembly complexes/intermediates the state of RNA folding in the vicinity of a specific site on a single small subunit ribosomal protein can be examined. Thus, the "dynamics" of protein-induced RNA conformational changes can be mapped. The complete set of plasmids encoding S1-S21 allow, for the first time, comprehensive mapping of RNA folding during assembly.
2) Identification of Non-Ribosomal Assembly Factors
Unlike advances made toward understanding the eukaryotic ribosome biogenesis machinery, where a multitude of assembly factors both protein and RNA have been identified, there is a lack of information regarding factors involved in prokaryotic ribosome assembly. A wealth of information indicates that such factors should be present in the prokaryotic system. Classical biochemical techniques, along with the recombinant protein reconstitution system, will be used to identify and characterize non-ribosomal assembly factors. Results from our group indicate that such factors can be identified and studied in E. coli. In vitro, as well as in vivo, studies will identify points of action of these factors. We are using affinity chromatography to purify assembly complexes formed in vivo to identify factors involved in this process. These approaches in combination with dynamic probing experiments will reveal events involved in ribosome biogenesis.
3) Identification of 16S rRNA nucleotides critical for assembly
Modification interference studies reveal specific nucleotides of 16S rRNA that are critical for different steps in assembly. Chemical probes are used to modify 16S rRNA in these assembly intermediates. We expect that some modifications allow continued assembly while others disrupt further assembly. Exploring modification-dependent assembly will enable us to address RNA folding and structure in combination with RNA-protein interactions.
4) In vivo roles for the ribosomal proteins
Although in recent years many have come to view the ribosomal proteins merely as scaffolds for RNA structure, more and more evidence indicates that the ribosomal proteins play discrete, functional roles in vivo. A complete set of plasmids encoding the small subunit ribosomal protein genes allows, for the first time, a comprehensive dominant negative screen. This screen, along with suppressor analysis, will help to elucidate the roles performed by these proteins and perhaps suggest explanations for ribosome complexity and conservation. We have undertaken genetic studies to study strains that bear deletions or specific mutations in ribosomal proteins to understand their functional roles in both ribosome biogenesis and function. The initial in vivo studies are complimented by in vitro biochemical experiments to further delineate the roles for these proteins.
5) Processing and Maturation of 16S rRNA
16S rRNA is transcribed with a long leader and trailer as part of a polycistron RNA that contains other rRNAs and transfer RNAs (tRNAs). The maturation process of rRNA is of great interest and importance. The initial excision of pre-16S and pre-23S rRNAs from the transcript has been well characterized. Although it is believed that there are multiple steps required to generate mature, functional rRNA very little is known about this process. The recombinant reconstitution system, genetics, and classical biochemistry will be used to study this RNA processing pathway.
Ultimately our goal is to understand how E. coli ribosomes are formed and how the biogenesis pathway is linked to function. Our studies will aid in our understanding of RNP assembly in general and may result in the identification of novel targets for antimicrobials.

Research Interests
- Macromolecular assembly
- RNA-protein recognition
- Dynamics of RNA folding
Selected Publications
- Calidas, D. and Culver, G.M. 2011. Interdependencies govern multidomain architecture in ribosomal small subunit assembly. RNA, 17:263-77. PMID: 21156960.
- Desai, P.M., Culver, G.M., and Rife, J.P. 2011. Site-directed mutants of 16S rRNA reveal important RNA domains for KsgA function and 30S subunit assembly. Biochemistry, 50:854-63. PMID: 21142019.
- Xu, Z. and Culver, G.M. 2010. Differential assembly of 16S rRNA domains during 30S subunit formation. RNA, 16:1990-2001. PMID: 20736336.
- Roy-Chauduri, B., Kirthi, N. and Culver, G.M. 2010. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc. Natl Acad Sci. 107:4567-72.
- Xu, Z. and Culver, G.M. 2009. Chemical probing of RNA and RNA/protein complexes. Methods of Enzymol. “Structural and Dynamic Methods in RNA Research”468:147-165.
- Pulicherla, N., Pogorzala, L.A., Xu, Z., O'Farrell, H.C., Musayev, F.N., Scarsdale, J.N., Sia, E.A., Culver, G.M. and Rife, J.P. 2009. Structural and functional divergence within the Dim1/KsgA family of rRNA methyltransferases.J Mol Biol. [Epub ahead of print] PMID: 19520088.
- Connolly, K., and Culver, G.M. 2009. Deconstructing Ribosome Construction.TiBS34:256-263.
- Connolly, K., J.P. Rife and Culver, G.M. 2008. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA.Mol Micro. 70:1062-1075.
- O'Farrell, H.C., Xu, Z., Culver, G.M, and Rife, J.P. 2008. Sequence and structural evolution of the KsgA/Dim1 methyltransferase family.BMC Res. Notes 1, 108.
- Xu, Z., O'Farrell, H.C., Rife, J.P., and Culver, G.M. 2008. A conserved rRNA methyltransferase regulates ribosome biogenesis.Nat Struct Mol Biol. 15:534-536.
- Roy-Chauduri, B., Kirthi, N., Kelley, T. and Culver, G.M. 2008. Suppression of a cold-sensitive defect in ribosomal protein S5 reveals a role for RimJ in ribosome biogenesis.Mol. Micro.68:1547-59.
- Lee, J.H., Culver, G., Carpenter, S., and Dobbs, D. 2008. Analysis of the EIAV Rev-responsive element (RRE) reveals a conserved RNA motif required for high affinity Rev binding in both HIV-1 and EIAV. PLoS ONE3:e2272.
- Culver, G.M. and Kirthi, N. 31 January 2008, posting date. Chapter 2.5.3, Assembly of the 30S ribosomal subunit. In A. Böck, R. Curtiss III, J. B. Kaper, F. C. Neidhardt, T. Nyström, K. E. Rudd, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. [Online.] ASM Press, Washington, D.C.
- Rife, J.P and Culver, G.M. 2007. Breaking the Cycle of Translation.Mol. Cell28:517-519.
- Dutcã, L.M. and Culver, G.M. 2007. Assembly of the 5' and 3' minor domains of 16S rRNA as monitored by tethered probing from ribosomal protein S20. J Mol Biol.376:92-108.
- 2007. Temperature-dependent RNP conformational rearrangements: analysis of binary complexes of primary binding proteins with 16S rRNA.J. Mol. Biol. 368:853-869.
- 2006. Primary ribosomal binding protein S15 plays a role in ribosome assembly and subunit association however it is not essential for E. coli viability.RNA12:1229-1239.
- 2006. A novel single amino acid change in small subunit ribosomal protein S5 has profound effects on translational fidelity..RNA12:2080-2091.
- 2005. Recent Developments in Factor-facilitated Ribosome Assembly.Methods36:313-320.
- 2005. Analysis of conformational changes in 16S rRNA during the course of 30S subunit assembly.J. Mol. Biol.354:340-357.
- 2004. Mapping structural differences between 30S ribosomal subunit assembly intermediates.Nat. Struct. Mol. Bio.11:179-186.
- 2004. Ribosomal protein-dependent orientation of the16 S rRNA environment of S15.J. Mol. Bio.335:1173-1185.
- 2004. Assembly of the 30S ribosomal subunit: Positioning ribosomal protein S13 in the S7 Assembly Branch.RNA10:1861-1866.
- 2003. Assembly of the 30S ribosomal subunit.Biopolymers68:234-249.
- 2003. Assembly of the central domain of the 30S ribosomal subunit: roles for the primary binding ribosomal proteins S15 and S8. J Mol Biol.330:373-383.
- 2003. Ribosomal proteins S21 and S13 function as control elements for translocation of the mRNA:tRNA complex.Mol. Cell12:321-328.
- 2003. Demonstration of the Role of the DnaK Chaperone System in Assembly of 30S Ribosomal Subunits Using a Purified in vitro System.RNA9:1418-1421.
- 2002. The DnaK chaperone system facilitates 30S ribosomal subunit assembly.Mol Cell.10:129-138.
- 2002. Sno-capped: 5' ends of preribosomal RNAs are decorated with a U3 SnoRNP.Chem Biol.9:777-779.
- 2001. Meanderings of the mRNA through the Ribosome.Structure (Camb)9:751-758.
- 2000. The location of protein S8 and surrounding elements of 16S rRNA in the 70S ribosome from combined use of directed hydroxyl radical probing and X-ray crystallography.RNA6:717-729.
- 2000. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes.Methods Enzymol318:461-475.
- 2000. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins.Methods Enzymol318:446-460.
- 1999. Identification of an RNA-Protein Bridge Spanning the Ribosomal Subunit Interface.Science285:2133-2135.
