
Patrick J. Murphy
Assistant Professor
PhD
- Office Location
- 2-9619 KMRB
- Telephone
- (585) 275-0113
- Web Address
- Website
Office Hours: By appointment
Research Overview
Our lab investigates the mechanisms that activate or silence genes. As cells divide during development, changes in gene expression provide each cell type with a specific identity and function. In a similar sense, when the gene expression patterns of normal adult cells change inappropriately, the cell identity also changes, and this can lead to carcinogenesis. We utilize the zebrafish model in combination with mammalian cell lines to investigate how epigenetic marks control gene expression patterns to drive cell state transitions. This work relies on classic genetics and developmental biology methods, new DNA sequencing technologies, and bioinformatics applications.
Zebrafish Genomics
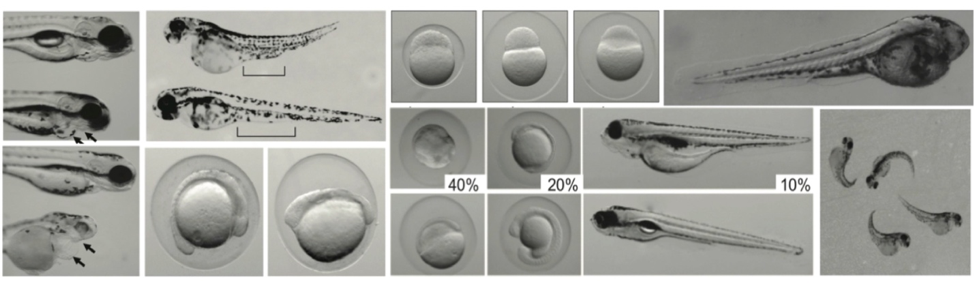
The zebrafish model has emerged as incredibly valuable when combined with genomics for understanding mechanisms of vertebrate developmental processes in vivo. Zebrafish fertilization and early organismal development occur ex utero, and each breeding pair can produce over 200 offspring in precisely timed matings. Therefore, many thousands of cells can be harvested at specific developmental stages for complex genomics studies.

The Murphy Lab
Research Interests
- Regulation of vertebrate gene transcription in vivo
- Epigenetics and chromatin
- Zebrafish genetics and genomics
- Developmental cell state transitions
- Bioinformatics
Selected Publications
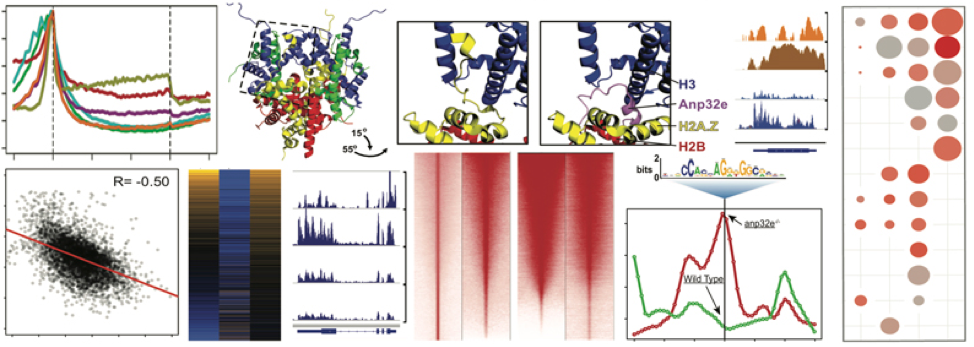
- Stefanelli G, Makowski CE, Brimble MA, Hall M, Reda A, Creighton SD, Leonetti AM, McLean TAB, Zakaria JM, Baumbach J, Greer CB, Davidoff AM, Walters BJ, Murphy PJ, Zovkic IB. The histone chaperone Anp32e regulates memory formation, transcription, and dendritic morphology by regulating steady-state H2A.Z binding in neurons. Cell Reports 2021; 36, 109551. PubMed PMID: 34407406.
- Meng FW, and Murphy PJ. Rolling uphill: in vivo reacquisition of pluripotency during cranial neural crest differentiation. Commun Biol 2021; 4, 626. PubMed PMID: 34017055.
- Murphy KE, Meng FW, Makowski CE, and Murphy PJ. Genome-wide chromatin accessibility is restricted by ANP32E. Nat. Commun. 2020 Oct 8;11, 5063. doi: 10.1038/s41467-020-18821-x. PubMed PMID: 33033242.
- Murphy PJ, Guo J, Jenkins TG, James ER, Hoidal JR, Huecksteadt T, Broberg DS, Hotaling JM, Alonso DF, Carrell DT, Cairns BR, Aston KI. NRF2 loss recapitulates heritable impacts of paternal cigarette smoke exposure. PLoS Genet. 2020 Jun 10;16(6):e1008756. doi: 10.1371/journal.pgen.1008756. PubMed PMID: 32520939.
- Rougeot J, Chrispijn ND, Aben M, Elurbe DM, Andralojc KM, Murphy PJ, Jansen PWTC, Vermeulen M, Cairns BR, Kamminga LM. Maintenance of spatial gene expression by Polycomb-mediated repression after formation of a vertebrate body plan. Development. 2019 Sep 30;146(19). doi: 10.1242/dev.178590. PubMed PMID: 31488564.
- Murphy PJ, Wu SF, James CR, Wike CL, Cairns BR. Placeholder Nucleosomes Underlie Germline-to-Embryo DNA Methylation Reprogramming. Cell. 2018 Feb 22;172(5):993-1006.e13. doi: 10.1016/j.cell.2018.01.022. Epub 2018 Feb 15. PubMed PMID: 29456083.
- Jenkins TG, James ER, Alonso DF, Hoidal JR, Murphy PJ, Hotaling JM, Cairns BR, Carrell DT, Aston KI. Cigarette smoking significantly alters sperm DNA methylation patterns. Andrology. 2017 Nov;5(6):1089-1099. doi: 10.1111/andr.12416. Epub 2017 Sep 26. PubMed PMID: 28950428.
- Guo J, Grow EJ, Yi C, Mlcochova H, Maher GJ, Lindskog C, Murphy PJ, Wike CL, Carrell DT, Goriely A, Hotaling JM, Cairns BR. Chromatin and Single-Cell RNA-Seq Profiling Reveal Dynamic Signaling and Metabolic Transitions during Human Spermatogonial Stem Cell Development. Cell Stem Cell. 2017 Oct 5;21(4):533-546.e6. doi: 10.1016/j.stem.2017.09.003. PubMed PMID: 28985528.
- Murphy PJ, Cairns BR. Genome-wide DNA methylation profiling in zebrafish. Methods Cell Biol. 2016 Jun 21;135:345-59. doi: 10.1016/bs.mcb.2016.05.002. PubMed PMID: 27443935.
- Murphy KE, Murphy PJ, Carrell DT. Epigenetic Changes in the Paternal Germline. In: Transgenerational epigenetics. Tollefsbol T, editor. Amsterdam ; Boston: Elsevier/Academic Press; 2014. Chapter 5.
- Murphy PJ, Cipriany BR, Wallin CB, Ju CY, Szeto K, Hagarman JA, Benitez JJ, Craighead HG, Soloway PD. Single-molecule analysis of combinatorial epigenomic states in normal and tumor cells. Proc Natl Acad Sci. 2013 May 7;110(19):7772-7. doi: 10.1073/pnas.1218495110. Epub 2013 Apr 22. PubMed PMID: 23610441.
- Benítez JJ, Topolancik J, Tian HC, Wallin CB, Latulippe DR, Szeto K, Murphy PJ, Cipriany BR, Levy SL, Soloway PD, Craighead HG. Microfluidic extraction, stretching and analysis of human chromosomal DNA from single cells. Lab Chip. 2012 Nov 21;12(22):4848-54. doi: 10.1039/c2lc40955k. PubMed PMID: 23018789.
- Cipriany BR, Murphy PJ, Hagarman JA, Cerf A, Latulippe D, Levy SL, Benítez JJ, Tan CP, Topolancik J, Soloway PD, Craighead HG. Real-time analysis and selection of methylated DNA by fluorescence-activated single molecule sorting in a nanofluidic channel. Proc Natl Acad Sci. 2012 May 29;109(22):8477-82. doi: 10.1073/pnas.1117549109. Epub 2012 May 14. PubMed PMID: 22586076.
- Park YJ, Herman H, Gao Y, Lindroth AM, Hu BY, Murphy PJ, Putnam JR, Soloway PD. Sequences sufficient for programming imprinted germline DNA methylation defined. PLoS One. 2012;7(3):e33024. doi: 10.1371/journal.pone.0033024. Epub 2012 Mar 5. PubMed PMID: 22403732.
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, Gotoh K, Hiura H, Arima T, Fujiyama A, Sado T, Shibata T, Nakano T, Lin H, Ichiyanagi K, Soloway PD, Sasaki H. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011 May 13;332(6031):848-52. doi: 10.1126/science.1203919. PubMed PMID: 21566194.
- Cipriany BR, Zhao R, Murphy PJ, Levy SL, Tan CP, Craighead HG, Soloway PD. Single molecule epigenetic analysis in a nanofluidic channel. Anal Chem. 2010 Mar 15;82(6):2480-7. doi: 10.1021/ac9028642. PubMed PMID: 20184350.
