
Amanda Larracuente
she/her/hers
Nathaniel and Helen Wisch Professor of Biology
Professor of Biology
Interim Department Chair
Research Active
Now accepting:
PhD students
Please email with inquiries.
Undergraduate researchers
Please email with inquiries.
- Office Location
- 343 Hutchison
- Telephone
- (585) 273-1693
- Web Address
- Website
Office Hours: By appointment
Research Overview
What do transposable elements, satellite DNAs and meiotic drive systems have in common? They are all selfish DNAs and can spread in genomes and populations without offering any benefit, and many times even causing harm, to their hosts. Our lab integrates genomic, cytological and molecular approaches to study selfish DNA and its impact on genome evolution. Our primary interest is in satellite DNA (repetitive DNA typically found at centromeres and telomeres) and meiotic drive. Lab projects focus on four related areas:
I. The functional and evolutionary genomics of satellite DNAs
We use genomic, cytological and molecular approaches to study the functional genomics of satellite DNAs and their dynamic evolution across taxa.
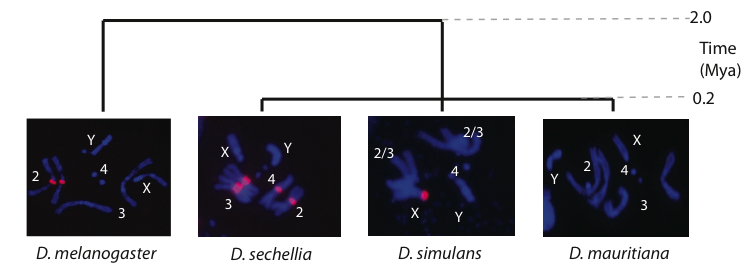
Rapid evolution of the Responder (Rsp) satellite: The Rsp satellite shows dynamic evolution over short evolutionary time periods, like many other satellites. Rsp is unique in D. melanogaster because it is a target of the meiotic drive complex, Segregation Distorter (SD). Figure from Larracuente, 2014.
II. Molecular mechanisms of meiotic drive
Meiotic drivers gain a transmission advantage through the germline. We use genetic, genomic and cytological approaches to determine the molecular mechanism of different drive systems. Our primary focus is on the selfish Segregation Distorter complex of D. melanogaster.
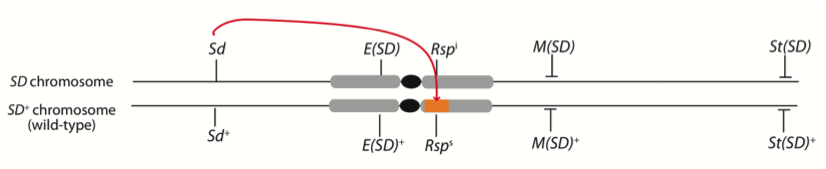
Organization of a Segregation Distorter (SD)chromosome: SD is a selfish gene complex on D. melanogaster chromosome 2. Males heterozygous for SD and a wild type chromosome transmit SD to >95% of their progeny. SD kills sperm with sensitive alleles of its target, the Responder (Rsp)satellite repeat, on chromosome 2R. We use genomic, cytological and molecular methods to study how SD kills Rspssperm.
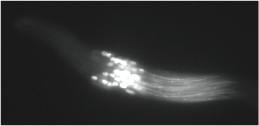
Sperm killing in Drosophila affinis:Sex ratio meiotic drivers kill Y-bearing sperm after meiosis in D. affinis. This figure shows an example of one phenotype in XSR2/Y testes. Approximately half of the nuclei in this sperm bundle are not properly condensed and will not make functional sperm.
III. Y chromosome evolution
Y chromosomes specialize in male fertility, but are generally gene-poor and dense in repetitive elements such as satellite DNAs. Differences in repeat content between Y chromosomes may impact genome evolution. We use population genomic and molecular methods to study Y chromosome evolution across various Drosophila species.
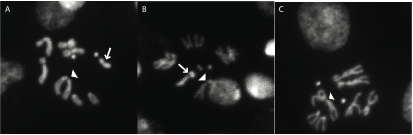
Y variation: Species of the obscura group segregate for Y chromosomes with different morphologies. This figure shows some of the different Y chromosomes in D. affinis. The arrows point to the Y chromosome and the arrowheads point to X chromosomes. Some D. affinis males lack a Y chromosome and are still fertile (panel C)!
IV. Centromere organization and evolution
Centromeres are essential chromosomal regions required for proper chromosome segregation during cell division. Despite their essential function, centromeres evolve rapidly. We combine comparative (epi)genomic and cytogenetic approaches in Drosophila species to study the evolution of centromere organization in Drosophila species.
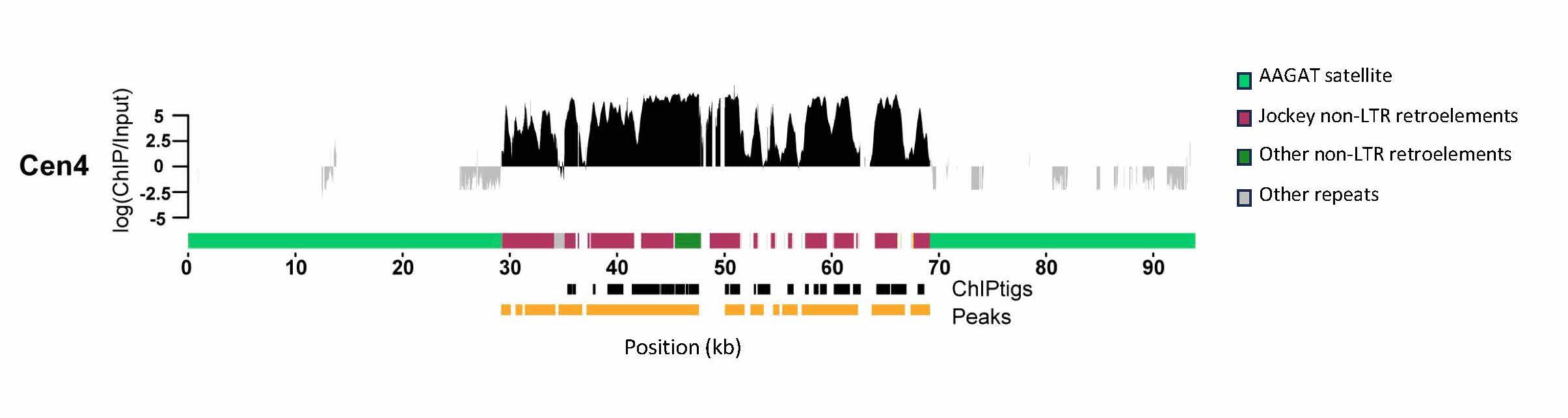
Drosophila melanogaster centromeres correspond to islands of retroelements in a sea of tandem satellite DNA. The figure shows the enrichment of the centromere-specific histone variant (CENP-A) ChIPseq reads on the retroelement island corresponding to the 4th chromosome centromere (figure from Chang, Chavan, Palladino, et al. 2019 PLOS Biology). We study how centromere organization varies within and across species to understand the role of DNA sequences in centromere evolution and function and the processes shaping their rapid evolution.
Research Interests
- Evolutionary genetics and genomics
- Intragenomic conflict and the evolution of selfish DNA
- Evolutionary and functional genomics of satellite DNA
- Sex chromosome and dot chromosome evolution in Drosophila
- Centromere organization and evolution
Selected Publications
- Courret, C. and A.M. Larracuente. 2023. High levels of intra-strain structural variation in Drosophila simulans X pericentric heterochromatin. GENETICS. iyad176. https://doi.org/10.1093/genetics/iyad176.
- Courret, C., Wei, X., and Larracuente, A.M. 2023. New perspectives on the causes and consequences of male meiotic drive. Curr Opinion Gen & Dev. https://doi.org/10.1016/j.gde.2023.102111.
- Martí, E. and A.M. Larracuente. 2023. Genetic conflict and the origin of multigene families: implications for sex chromosome evolution. Proc R. Soc. B. https://doi.org/10.1098/rspb.2023.1823.
- Navarro-Domínguez, B., Chang, C-H, Brand, C, Muirhead, C., Presgraves, D.C., and A.M. Larracuente. 2022 Epistatic selection on a selfish Segregation Distorter supergene: drive, recombination, and genetic load. eLife 2022;11:e78981 DOI: 10.7554/eLife.78981
- Chang, C-H., Gregory, L.E., Gordon, K.E., Meiklejohn, C.D., Larracuente, A.M. 2022. Unique structure and positive selection promote the rapid divergence of Drosophila Y chromosomes. eLife 2022;11:e75795 DOI: 10.7554/eLife.75795
- Wei,X., Eickbush,D.G., Speece,I., A.M. Larracuente. 2021. Heterochromatin-dependent transcription of satellite DNAs in the Drosophila melanogaster female germline. eLife 2021;10:e62375. doi: 10.7554/eLife.62375
- Sproul, J., Khost, D.E.*, Eickbush, D.G., Negm, S., Wei, X., Wong, I., and A.M. Larracuente 2020. Dynamic evolution of euchromatic satellites on the X chromosome in Drosophila melanogaster and the simulans clade. Molecular Biology and Evolution. 37(8)2241-2256, https://doi.org/10.1093/molbev/msaa078
- Chang, C-H*, Chavan, A*, Palladino, J.*, Wei, X., Martins, N.M.C., Santinello, B., Chen, C-C, Erceg, J., Beliveau, B.J., Wu, C-T, Larracuente, A.M., and Mellone, B.G. 2019. Islands of retroelements are major components of Drosophila melanogaster centromeres. PLoS Biology https://doi.org/10.1371/journal.pbio.3000241.
- Chang, C-H. and A.M. Larracuente. 2019. Heterochromatin-enriched assemblies reveal the sequence and organization of the Drosophila melanogaster Y chromosome. GENETICS 211: 333-348. doi: 10.1534/genetics.118.301765.
- Khost, D.E., D. Eickbush, A.M. Larracuente. 2017. Single molecule long read sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Research . http://www.genome.org/cgi/doi/10.1101/gr.213512.116.
- Chang, C-H. and A.M. Larracuente. 2017. Structural changes following the reversal of a Y chromosome to an autosome in Drosophila pseudoobscura. Evolution. 71-5:1285-1296. http://dx.doi.org/10.1111/evo.13229.
