
Dragony Fu
Mercer Brugler Distinguished Teaching Professor
Professor of Biology
Director, Graduate Affairs and Admissions in Biology
Research Active
Now accepting:
PhD students
Please email with inquiries.
- Office Location
- 333 Hutchison
- Telephone
- (585) 276-3346
- Web Address
- Website
Office Hours: By appointment
Research Overview
- Biological functions of RNA modifications
- Human disorders linked to RNA modifications
- Regulation of protein synthesis
Our laboratory investigates the emerging roles of RNA modification in biological processes ranging from protein synthesis to neurodevelopment. We use an integrated biochemical, molecular, and genetic approach that employs mammalian tissue culture cells, human brain organoids, and animal models such as Drosophila and mice. Through this approach, we have discovered novel targets and functions for RNA modification. The pathways and mechanisms identified through our studies have provided critical insight into multiple aspects of human health and disease, including anti-cancer chemotherapy, neurological disorders, and aging.
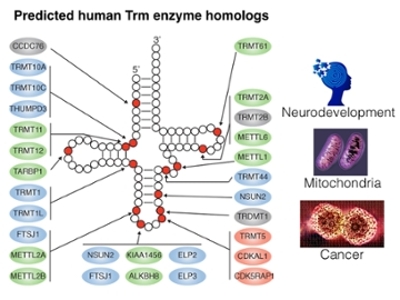
Research Interests
- Biological functions of RNA modifications
- Human disorders linked to RNA modifications
- Regulation of protein synthesis
Recent Publications
Caley Hogan, Scott Gratz, Jennifer L. Dumouchel, Rajan Thakur, Ambar Delgado Carrión, et al. Expanded tRNA methyltransferase family member TRMT9B regulates synaptic growth and function. EMBO reports, 2023 https://doi.org/10.15252/embr.202356808
Kejia Zhang, Patrick Eldin, Jessica H. Ciesla, Laurence Briant, Jenna M. Lentini, et al. Proteolytic cleavage and inactivation of the TRMT1 tRNA modification enzyme by SARS-CoV-2 main protease , 2023 https://doi.org/10.1101/2023.02.10.527147
Longan ER, Ramos J, Fu D ADATscan - A flexible tool for scanning exomes for wobble inosine-dependent codons reveals a neurological bias for genes enriched in such codons in humans and mice. microPublication biology, 2023 https://doi.org/10.17912/micropub.biology.000675
Jenna M. Lentini, Hessa S. Alsaif, Eissa Faqeih, Fowzan S. Alkuraya, Dragony Fu DALRD3 encodes a protein mutated in epileptic encephalopathy that targets arginine tRNAs for 3-methylcytosine modification Nature Communications, 2020 https://doi.org/10.1038/s41467-020-16321-6
Felix Hagelskamp, Kayla Borland, Jillian Ramos, Alan G Hendrick, Dragony Fu, et al. Broadly applicable oligonucleotide mass spectrometry for the analysis of RNA writers and erasers in vitro Nucleic Acids Research, 2020 https://doi.org/10.1093/nar/gkaa091
Kejia Zhang, Jenna M. Lentini, Christopher T. Prevost, Mais O. Hashem, Fowzan S. Alkuraya, et al. An intellectual disability‐associated missense variant in TRMT1 impairs tRNA modification and reconstitution of enzymatic activity Human Mutation, 2020 https://doi.org/10.1002/humu.23976
Biallelic variants in CTU2 cause DREAM-PL syndrome and impair thiolation of tRNA wobble U34. Human mutation, 2019 https://doi.org/10.1002/humu.23870
Jillian Ramos, Lu Han, Yan Li, Felix Hagelskamp, Stefanie M. Kellner, et al. Formation of tRNA Wobble Inosine in Humans Is Disrupted by a Millennia-Old Mutation Causing Intellectual Disability Molecular and Cellular Biology, 2019 https://doi.org/10.1128/MCB.00203-19
Jillian Ramos, Dragony Fu The emerging impact of tRNA modifications in the brain and nervous system Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, 2019 https://doi.org/10.1016/j.bbagrm.2018.11.007
Chen Gu, Jillian Ramos, Ulrike Begley, Peter C. Dedon, Dragony Fu, et al. Phosphorylation of human TRM9L integrates multiple stress-signaling pathways for tumor growth suppression Science Advances, 2018 https://doi.org/10.1126/sciadv.aas9184
For more, please visit my ORCID Profile
