![]()
The R2 insertion mechanism has been studied by expressing the R2 open reading frame in bacteria and characterizing the activities of the purified protein product. The single 120 kilodalton protein has been shown capable of conducting most of the steps of an integration reaction. The R2 protein is composed of three domains (see Figure): an N-terminal DNA binding domain (blue shading), a central reverse transcriptase domain (green shading) and a C-terminal DNA binding and endonuclease domain (red shading). The two DNA binding domains bind the target DNA sequences either upstream or downstream of the insertion site. These specific bindings allow precise cleavage of the two strands of the DNA target site. The R2 protein also has the ability to bind two different RNA sequences: RNA corresponding to the 3’ untranslated region of the R2 element and RNA near the 5’ end of the element. Binding of either of these RNAs determines whether the R2 protein binds the upstream or downstream DNA sequences. The reverse transcriptase encoded by R2 has a number of unusual characteristics. First it can use the 3’ end of a DNA strand to prime reverse transcription. No sequence complementarity is needed in this priming reaction. Second the polymerase can jump from the 5’ end of one RNA or DNA template to the 3’ end of another RNA or DNA template again in the absence of sequence identity between the two templates (end-to-end template jumping). Finally the polymerase is highly processive (i.e. the length a enzyme can polymerase without falling off the template) on both RNA and DNA templates. During this polymerization the enzyme can displace a DNA or RNA strand annealed to the template.
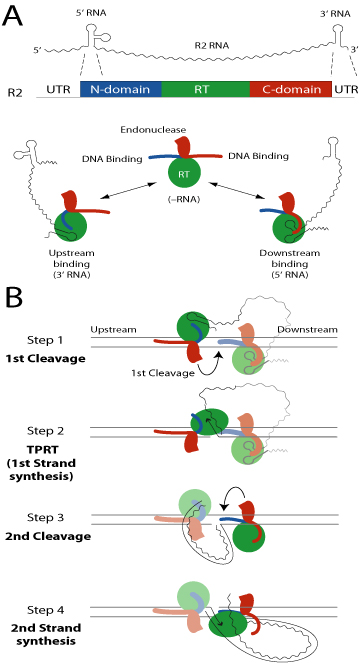
Model of R2 retrotransposition. (A) Domain structure of the R2 protein and the conformational changes involved
in the binding of the 5’ or 3’ ends of the RNA transcript. (B) The four steps of a complete R2 integration reaction.
Based on these properties of the R2 protein our current model of R2 retrotransposition is as follows. R2 protein bound to the 3' UTR RNA sequesters the N-terminal domain, exposing only the C-terminal DNA binding domain for binding upstream of the insertion site. R2 protein bound to the 5' RNA sequesters the C-terminal DNA binding domain, exposing only the N-terminal DNA binding domain to bind the downstream site. R2 retrotransposition is proposed to be catalyzed by these two subunits in four steps. (Step 1) the endonuclease (red oval) from the upstream subunit is responsible for first strand cleavage. (Step 2) The RT (green oval) of the upstream subunit catalyzes reverse transcription of the RNA template using the cleaved DNA target site as primer, a reaction we call Target Primed Reverse Transcription, TPRT. (Step 3) The downstream subunit cleaves the second DNA strand. (Step 4) The downstream subunit provides the polymerase to perform second strand TPRT displacing the RNA template as it uses the first DNA strand as template.
|
Home | Research overview | Mech of retrotransposition | Evolution of the rDNA locus |Regulation of activity| Publications | Members | |