![]()
Evolution of the rDNA locus
While theoretical and population genetic studies have attempted to explain the concerted evolution of rDNA loci, few studies have followed specific changes in the structure of these loci over time. Only by such direct observations will we be able to understand the recombinational mechanisms that give rise to the concerted evolution of the locus. We are conducting detailed studies of the rDNA locus of several Drosophila species using both nucleotide sequence variation and R1 and R2 insertions as specific markers. One study is evaluating the short-term evolutionary dynamics of the rDNA locus by comparing the rDNA loci in replicate Drosophila melanogaster lines after 400 generations. Hundreds of new R1 and R2 insertions and deletions were scored in these lines. The total number of rDNA units on either the X or Y chromosome of each line varied nearly three-fold, as did the fraction of units inserted with R1 and R2 (see Figure). The variation in total number of units and fraction occupied by insertions in the replicate lines were comparable to that found in natural population surveys. These studies have given rise to a BAC cloning project to recover significant portions of the rDNA loci of one of the lines. With a map of the distribution of variation along large segments of the rDNA locus from this line, we will be able to use the variation scored in all replicate lines to provide new insights into the relentless forces tht rapidly shape the landscape of the rDNA locus.
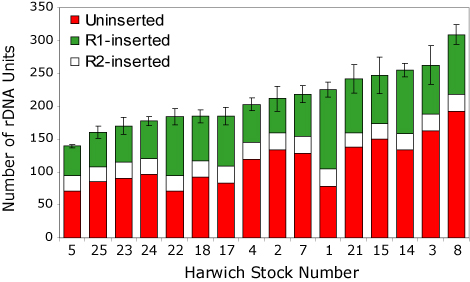
Variation in the X chromosome rDNA locus size and load of R1 and R2 elements in 16 D. melanogaster replicate lines reared separately for 400 generations. The total number of units in each line is indicated by the height of each bar. The number of uninserted, R1 inserted and R2 inserted units are indicated by the shading within the bar.
Another study utilizes the data from whole genome shot-gun sequencing efforts in multiple species of Drosophila to follow the longer term cumulative effects of recombination and element activity on the structure of the rDNA locus. The advent of genomic sequence data introduces a new era in our attempts to understand the effects of recombinations and retrotranspositions on the evolution of the rDNA locus. We have developed a comprehensive, rapid means to identify sequence polymorphisms within the rDNA units of any genome. These studies are providing important clues to how nucleotide sequence changes spread through the rDNA loci as they become either fixed or eliminated. In addition, these studies provide insights on the frequency and effects of R1 and R2 insertions.
Home | Research overview | Mech of retrotransposition | Evolution of the rDNA locus |Regulation of acitivity| Publications | Members | |