Wolbachia Research
Our research on Wolbachia spans many different areas from genome evolution, host-Wolbachia interactions, the role of Wolbachia in host reproductive isolation, and Wolbachia-host lateral gene transfer. An introduction to Wolbachia biology can be found here or see Werren et al. 2009.
Below is a summary of some recent research projects:
A mitochondrial-Wolbachia sweep in N. vitripennis in North America
Our recent work on mitochondrial genetic variation suggests that a mitochondrial-Wolbachia sweep in has occurred in North America. We have compared levels of genetic variation in European and North American samples of N. vitripennis. European N. vitripennis populations have 7-fold higher sequence variation in their mitochondrial sequence compared to North American samples but similar levels of microsatellite and nuclear sequence variation. Variation in the North American mitochondria is extremely low (π=0.31%), despite a highly elevated mutation rate (~35-40 times higher than the nuclear genes). The data are indicative of a mitochondrial sweep in the North American populations, possibly due to Wolbachia infections which are maternally co-inherited with the mitochondria. Because of similar levels of nuclear variation, the data could not resolve whether N. vitripennis originated in the New or the Old World (Raychoudhury et al. 2010).
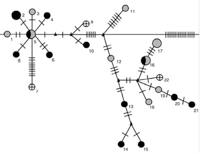
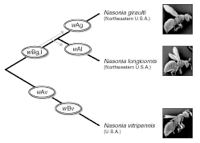
Modes of Acquisition of Wolbachia in Nasonia
One of the key features of Wolbachia biology is its ability to move within and between host species, which contributes to the impressive diversity and range of infected hosts. We have characterized the diversity and modes of movement of Wolbachia within the wasp genus Nasonia. Eleven different Wolbachia were found in the four species of Nasonia, including five newly identified infections. Five infections were acquired by horizontal transmission from other insect taxa, three have been acquired by hybridization between two Nasonia species, which resulted in a mitochondrial-Wolbachia sweep from one species to the other, and at least three have co-diverged during speciation of their hosts. The results show that a variety of transfer mechanisms of Wolbachia are possible even within a single host genus (Raychoudhury et al. 2009).
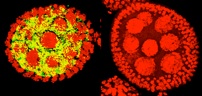
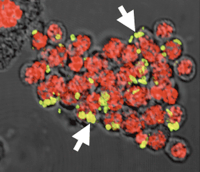
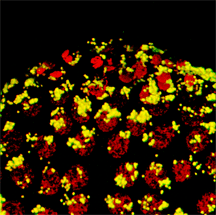
Wolbachia, Cytoplasmic Incompatibility and Spermatogenesis
A study of spermatogenesis in the parasitic wasp Nasonia vitripennis reveals that Wolbachia are not required in individual spermatocytes or spermatids to modify sperm. In N. vitripennis, Wolbachia modify nearly all sperm, but are found only in approximately 28% of developing sperm, and are also found in surrounding cyst and sheath cells. In the beetle Chelymorpha alternans, Wolbachia can modify up to 90% of sperm, but were never observed within the developing sperm or within the surrounding cyst cells; they were abundant within the outer testis sheath. We conclude that the residence within a developing sperm is not a prerequisite for Wolbachia-induced sperm modification, suggesting that Wolbachia modification of sperm may occur across multiple tissue membranes or act upstream of spermiogenesis. (Clark et al. 2008)
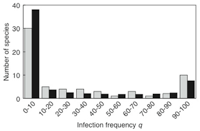
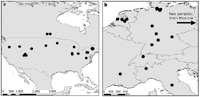
How many species are infected with Wolbachia? Answer: 66%
Estimates suggest that at least 20% of all insect species are infected with Wolbachia. These estimates result from several Wolbachia screenings in which numerous species were tested for infection; however, tests were mostly performed on only one to two individuals per species. The actual percent of species infected will depend on the distribution of infection frequencies among species. We present a meta-analysis that estimates percentage of infected species based on data on the distribution of infection levels among species. We used a beta-binomial model that describes the distribution of infection frequencies of Wolbachia, shedding light on the overall infection rate as well as on the infection frequency within species. Our main findings are that (1) the proportion of Wolbachia-infected species is estimated to be 66%, and that (2) within species the infection frequency follows a 'most-or-few' infection pattern in a sense that the Wolbachia infection frequency within one species is typically either very high (>90%) or very low (<10%) (Hilgenboecker et al. 2008).
Lateral gene transfer
Until recently, the transfer of genes from bacteria to multicellular eukaryotes was considered very rare. Our recent work with Wolbachia however has shown that such lateral gene transfers are widespread. Wolbachia genes have been transferred to the chromosomes of hosts at relatively high frequencies. In the fruitfly, Drosophila ananassae, nearly the entire Wolbachia chromosome has been integrated into one of the fly chromosomes (Dunning Hotopp et al 2007). For a stories about this work see here and here.
Bidirectional incompatibility in Nasonia
The parasitic wasp species complex Nasonia (N. vitripennis, N. longicornis and N. giraulti) harbor at least six different Wolbachia that cause CI. Each species have double infections with a representative from both the A and B Wolbachia subgroups. CI relationships of the A and B Wolbachia of N. longicornis with those of N. giraulti and N. vitripennis are investigated here. We demonstrate that all pairwise crosses between the divergent A strains are bidirectionally incompatible. We were unable to characterize incompatibility between the B Wolbachia, but we establish that the B strain of N. longicornis induces no or very weak CI in comparison to the closely related B strain in N. giraulti that expresses complete CI. Taken together with previous studies, we show that independent acquisition of divergent A Wolbachia has resulted in three mutually incompatible strains, whereas codivergence of B Wolbachia in N. longicornis and N. giraulti is associated with differences in CI level. Understanding the diversity and evolution of new incompatibility strains will contribute to a fuller understanding of Wolbachia invasion dynamics and Wolbachia-assisted speciation in certain groups of insects (Bordenstein and Werren 2007).
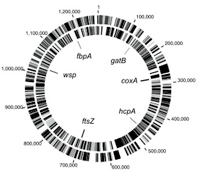
A Multilocus sequence typing system Wolbachia
We have developed a multilocus sequence typing (MLST) scheme as a universal genotyping tool for Wolbachia. Internal fragments of five ubiquitous genes (gatB, coxA, hcpA, fbpA, and ftsZ) were chosen, and primers that amplified across the major Wolbachia supergroups found in arthropods, as well as other divergent lineages, were designed. A supplemental typing system using the hypervariable regions of the Wolbachia surface protein (WSP) was also developed. Thirty-seven strains belonging to supergroups A, B, D, and F obtained from singly infected hosts were characterized by using MLST and WSP. The number of alleles per MLST locus ranged from 25 to 31, and the average levels of genetic diversity among alleles were 6.5% to 9.2%. A total of 35 unique allelic profiles were found. The results confirmed that there is a high level of recombination in chromosomal genes. MLST was shown to be effective for detecting diversity among strains within a single host species, as well as for identifying closely related strains found in different arthropod hosts. Identical or similar allelic profiles were obtained for strains harbored by different insect species and causing distinct reproductive phenotypes. Strains with similar WSP sequences can have very different MLST allelic profiles and vice versa, indicating the importance of the MLST approach for strain identification. The MLST system provides a universal and unambiguous tool for strain typing, population genetics, and molecular evolutionary studies (Baldo et al. 2006).