Nasonia Research
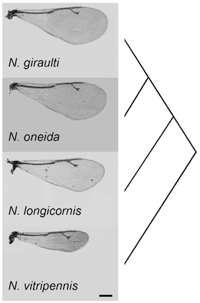
Nasonia wasps, commonly known as jewel wasps, consist of four closely related species, N. vitripennis, N. giraulti, N. longicornis, and N. oneida (Darling and Werren 1990, Raychoudhury et al 2008). These small insects, about 1/4 the size of Drosophila melanogaster, sting and lay their eggs upon the pupae of various flies (e.g. blow flies, flesh flies and house flies). Nasonia are very easy to work with. They have a short generation time (2 weeks at 25C) and large family sizes. Individuals are easily sexed in the immobile pupal stage, adults do not readily fly, allowing handling without anesthetization. Pupae and adults are cold-hardy, allowing storage under refrigeration for several weeks, and larvae have an inducible overwintering diapause, which permits strain maintenance under refrigeration for ~18 months.
A key feature is their form of sex determination, called haplodiploidy, which makes them particularly suited for genetic studies. Females are diploid and develop from fertilized eggs, whereas males are haploid and develop parthenogenetically from unfertilized eggs. This form of sex determination allows researchers to exploit many of the advantages of haploid genetics in an otherwise complex eukaryotic organism. As a result, Nasonia are emerging as models for studies of complex genetic traits, because such traits are readily dissected genetically by taking advantage of male haploidy.
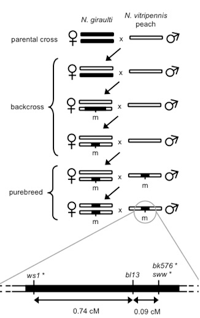
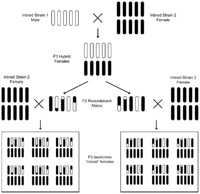
Normally, the three Nasonia species are genetically isolated from each other due to Wolbachia, an intracellular bacterium that causes sperm-egg incompatibilities (Werren 1997). However, these bacteria are easily eliminated by antibiotics – the species are then interfertile and genes and genetic regions can readily be moved between them by backcrossing (e.g. Weston et al 1999). This allows efficient fine-scale mapping and cloning of genes involved in species differences, making Nasonia an excellent model for evolutionary genetics.
Nasonia can be considered the “lab rat” of the Hymenoptera. In fact, it has inherent advantages as a genetic system that rivals even the current model insect systems that have been much more developed. It uniquely combines in a complex eukaryotic system the features of haploid genetics, short generation time, laboratory tractability, and availability of closely related inter-fertile species. Other important resources and tools include genome sequences from 5 closely related species, extensive expression array data, genotyping microarrays, and systemic RNAi.
Availability of genome sequences from 3 closely related species has greatly accelerated fine-scale mapping and cloning of QTL. Interfertile species and male haploidy have been used to rapidly map genome features onto the five chromosomes of Nasonia (Niehuis et al 2010). Two additional species (N. oneida and Trichomalopsis sarcophagae) have recently been sequenced and assembly. Analysis of these genomes is underway. More on genomics of Nasonia are available through the link to the right.
Our research on Nasonia is focused on evolutionary genetics, and spans and integrates many different research areas, including genetics, genomics, behavioral ecology, molecular evolution, and development.
Below is a summary of some recent publications
The Parasitoid Wasp Nasonia: An Emerging Model System With Haploid Male Genetics
Nasonia has several features that make it an excellent genetic system, including a short generation time, ease of rearing, interfertile species, visible and molecular markers, and a sequenced genome. The form of sex determination, called "haplodiploidy," makes Nasonia particularly suitable as a genetic tool. This allows geneticists to exploit many of the advantages of haploid genetics in an otherwise complex eukaryotic organism. Nasonia readily inbreeds, permitting production of isogenic lines, and the four species in the genus are interfertile (after removal of the endosymbiont Wolbachia), facilitating movement of genes between the species for efficient positional cloning of quantitative trait loci (QTL). Genome sequencing of the genetic model Nasonia vitripennis and two interfertile species, Nasonia giraulti and Nasonia longicornis, is now completed. This genome project provides a wealth of interspecies polymorphisms (e.g., single nucleotide polymorphisms [SNPs], insertion-deletions [indels], microsatellites) to facilitate positional cloning of genes involved in species differences in behavior, morphology, and development. Advances in the genetics of this system also open a path for improvement of parasitoid insects as agents of pest control.
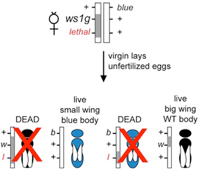
Wing size QTL in Nasonia
The genetic basis of morphological differences between species is still poorly understood. We investigated the genetic basis of a large change in male-specific wing size between two closely related species of Nasonia. A major genetic change behind this difference, called wing-size1 (ws1), increases male (but not female) wing size by 45%, altering both cell size and cell number. Using a genetic mapping approach, we determined that ws1 is located in non-protein-coding DNA region between the gene prospero (a transcription factor involved in neurogenesis) and the master sex-determining gene doublesex. Wing size reduction is correlated with an increase in doublesex expression level that is specific to developing male wings. This lends support to the hypothesis that such non-coding changes should be important in the evolution of body form because of the importance of non-coding DNA in regulating when and where genes are activated in development. This study demonstrates the feasibility of efficient positional cloning of quantitative trait loci (QTL) involved in a broad array of phenotypic differences among Nasonia species. (Loehlin et al. 2010)
The genetic basis of host preference in Nasonia
The genetic basis of host preference is relevant to important questions in evolutionary biology and agriculture. We have shown that a major locus strongly influences host preference in Nasonia. Nasonia consists of both a generalist and specialist species; N. vitripennis is a generalist that parasitizes a diverse set of fly hosts whereas the other Nasonia species specialize on Protocalliphora (bird blowflies). In laboratory choice experiments using Protocalliphora and Sarcophaga (flesh flies), N. vitripennis shows a preference for Sarcophaga while N. giraulti shows a preference for Protocalliphora. Through a series of interspecies crosses we have introgressed a major locus affecting host preference from N. giraulti into N. vitripennis. The N. giraulti allele is dominant and greatly increases preference for Protocalliphora pupae in the introgression line relative to the recessive N. vitripennis allele. This is the first introgression of the host preference of one parasitoid species into another, as well as one of the few cases of introgression of a behavioral gene between species (Desjardins et al. 2010).
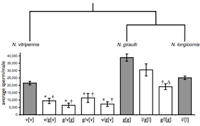
The evolution of wings size in Nasonia
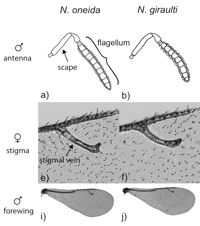
How do morphological differences between species evolve at the genetic level? The forewings of flightless N. vitripennis males are 2.3 times smaller than males of their flighted sister species N. giraulti. A major genetic contributor to this difference is the sex-specific widerwing (wdw) locus, which we have backcrossed from N. giraulti into N. vitripennis and mapped to an 0.9 megabase region of chromosome 1. This wdw introgression increases male but not female forewing width by 30% through wing region-specific cell size and cell number changes. Introgressing the same locus from the other species in the genus, N. longicornis and N. oneida, into N. vitripennis produces intermediate and large male wing sizes. To our knowledge, this is the first study to introgress a morphological quantitative trait locus (QTL) from multiple species into a common genetic background. Epistatic interactions between wdw and other QTL are also identified by introgressing wdw from N. vitripennis into N. giraulti. The main findings are 1) the changes at wdw have sex- and region-specific effects and could therefore be regulatory, 2) the wdw locus appears to be a co-regulator of cell size and cell number, and 3) the wdw locus has evolved different wing width effects in three species (Loehlin et al. 2010).
The effects of hybridization on male mating behavior and sperm production
Our recent work has show that hybrid males suffer two reproductive disadvantages, an inability to properly court females and decreased sperm production. While hybrids between the younger species pair (N. giraulti and N. longicornis) have nearly normal male mating behavior and sperm production, hybrids between the older species pair (N. giraulti and N. vitripennis) suffer from greatly decreased ability to mate as well as severe reductions in sperm production. This effect on hybrid males, lowered sperm counts rather than non-functional sperm, is different from most described cases of hybrid male sterility and may represent an earlier stage of hybrid sperm breakdown. The results add to previous studies of F2 hybrid inviability and behavioral sterility, and indicated that Wolbachia induced hybrid incompatibility has arisen early in species divergence, relative to behavioral sterility and spermatogenic infertility (Clark et al. 2010).
A New Species of Nasonia
We have recently discovered a new species, Nasonia oneida, distinguished by its behavioral, genetic and morphological features. It is isolated by both pre-mating and post-mating reproductive isolation with the other Nasonia species. N. oneida is closely related to N. giraulti. An important characteristic of N. oneida is the strong mate discrimination shown by the females against all the other Nasonia species (Raychoudhury et al. 2010).
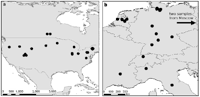
A mitochondrial-Wolbachia sweep in N. vitripennis in North America
Our recent work on mitochondrial genetic variation suggests that a mitochondrial-Wolbachia sweep in has occurred in North America. We have compared levels of genetic variation in European and North American samples of N. vitripennis. European N. vitripennis populations have 7-fold higher sequence variation in their mitochondrial sequence compared to North American samples but similar levels of microsatellite and nuclear sequence variation. Variation in the North American mitochondria is extremely low (π=0.31%), despite a highly elevated mutation rate (~35-40 times higher than the nuclear genes). The data are indicative of a mitochondrial sweep in the North American populations, possibly due to Wolbachia infections which are maternally co-inherited with the mitochondria. Because of similar levels of nuclear variation, the data could not resolve whether N. vitripennis originated in the New or the Old World (Raychoudhury et al. 2010).
Modes of Acquisition of Wolbachia in Nasonia
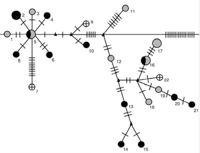
One of the key features of Wolbachia biology is its ability to move within and between host species, which contributes to the impressive diversity and range of infected hosts. We have characterized the diversity and modes of movement of Wolbachia within the wasp genus Nasonia. Eleven different Wolbachia were found in the four species of Nasonia, including five newly identified infections. Five infections were acquired by horizontal transmission from other insect taxa, three have been acquired by hybridization between two Nasonia species, which resulted in a mitochondrial-Wolbachia sweep from one species to the other, and at least three have co-diverged during speciation of their hosts. The results show that a variety of transfer mechanisms of Wolbachia are possible even within a single host genus (Raychoudhury et al. 2009).
.