Research
We are interested in the roles of basic cellular behaviors in embryonic patterning, especially in the context of RNA localization and asymmetric cell division. At the same time, we examine how the structures of developmental and cellular processes influence patterns of evolutionary change. We address these questions using the embryos of the snail Ilyanassa, which have particular advantages for the study of RNA localization, asymmetric cell division and the evolution of early embryonic patterning. [Ilyanassa as a model system]
The lab is mainly focused on two projects:
The role of centrosomal RNA localization in asymmetric cell division and embryonic patterning.
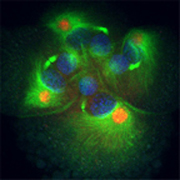
Asymmetric cell division is a crucial mode of cell fate specification in multicellular organisms, but it is relatively poorly understood. For instance, it is not yet clear whether there is a conserved mechanism of asymmetric cell division among metazoans. In the Ilyanassa embryo, almost all of the early cleavages are asymmetric, and the large size and accessibility of the embryo allow approaches that are not available in other systems. Thus, Ilyanassa is an important complement to existing models of asymmetric cell division.
Almost all of the early cell divisions in the Ilyanassa embryo are overtly asymmetric, and cell fates are specified during these early asymmetric cell divisions. However, until recently no putative determinant molecules had been identitified. We found that many RNAs are specifically segregated in these divisions, by a novel centrosome-dependent mechanism. This mechanism is related to a mechanism of subcellular localization that involved in oogenesis in animals as diverse as humans and flies. We have carried out a screen to find RNAs with specific segregation (Kingsley et al, 2007). This work shows that a large fraction of RNAs are specifially localized and segregated (3-4%), and that RNA segregation occurs in all asymmetric cell divisions in the early embryo. It also shows that the mechanisms of localization are intricately regulated, since the patterns of the 16 localized RNAs we characterized were all unique. We are using the localized RNAs we have recovered to address several basic questions.
How is the specificity and intricacy of RNA localization achieved? We are empirically determining the sequences and secondary structures that mediate localization, and using these to test how localization to different cells is regulated, both within and between RNAs. This work will have general implications for the regulation of RNA localization in development and oogenesis. It may also help explain how interative RNA localization events of the same RNA in successive cleavage cycles can define an embryonic lineage.
Are localized RNAs important for embryonic patterning? We have developed several new methods for Ilyanassa, including pressure injection and gene knockdown with morpholino oligos and siRNA. We are characterizing the developmental roles of several putative patterning genes. The first localized RNA we have characterized is the Ilyanassa Nanos mRNA, which is involved with the development of the endomesodermal blast cell lineage.
Is centrosomal localization required for the normal function of localized RNAs? We are using results from the above approaches to address whether the centrosomal localization we observe is a necessary part of of the RNA segregation process, and required for developmental roles of the localized RNAs. For instance, we have found that the same ~100 bp RNA sequence that can mediate specific centrosomal localization is also sufficient to mediate cortical localization and segregation, suggesting that these two steps are tightly linked.
Patterning the spiralian embryo
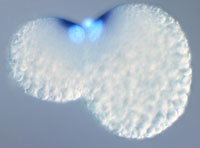
The process of early embryogenesis in animals involved dramatic cell biological and developmental phenomena. The evolution of this process during the early radiation of the animal kingdom generated much of the fundamental diversity that we can observe among living animal groups.
One compelling reason for studying Ilyanassa is that it is an excellent model for a conserved but enigmatic mode of early embryogenesis known as spiralian development. The advent of molecular systematics has dramatically reorganized the relationships of animal phyla in the last 15 years (reviewed in Halanych, 2004). One of the most striking findings of this work was that there are three large clades of phyla among bilaterian animals: the Dueterostomia, including chordates, urochordates, echinoderms, and hemichordates; the Ecdysozoa, including arthropods, nematodes and several other marine phyla; and the Lophotrochozoa, which includes the remaining ten or so animal phyla including molluscs, annelids, turbellarian flatworms, nemerteans, bryozoans and phoronids. The Lophotrochozoa is the largest of the three in terms of phyla, and is arguably the most diverse and disparate in terms of animal body plans. However, this group is the only one of the three superphyla that does not have a major model system for the study of development. Within the Lophotrochozoans, several phyla share a highly conserved mode of early development called spiralian development, which is characterized by similarlities in the cleavage pattern, the embryonic fate map, and the larval morphology. Since it is shared by several phyla, spiralian development is a good place to start in an attempt to understand the mechanisms of Lophotrochozoan development.
The Ilyanassa embryo has classic spiralian development. It also presents a number of practical and experimental advantages that make it an excellent model system for studying spiralian development. [Ilyanassa as a model system] We are using Ilyanassa to characterize the key patterning events of the spiralian embryo at the molecular level.
How is the secondary axis specified? Spiralian embryos initially have four-fold symmetry, but this is broken to generate a secondary axis and bilateral symmetry. This event involves the specification of the"D quadrant" lineage, either by localized determinants or by cell signaling. We are trying to understand how the D quadrant is specified in Ilyanassa, by studying a localized RNA that seems to be required for this process.
How are the quartet identities established? An elegant series of cell transplantation experiments have demonstrated that in the Ilyanassa embryo, the birth order of the micromere cells (aka quartet identity) determines the set of fates that they can adopt (Sweet, 1998). We are testing whether localized RNAs we recovered in our in situ screen (Kingsley et al, 2007) are involved in these specification events.
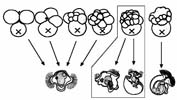
How does organizer signaling influence the pattern of micromere fates? After the 5th cleavage cycle, the D macromere cell (known as 3D) acts as an embryonic organizer to induce particular fates in the cells of the A, C and D lineages (Clement, 1962; Lambert and Nagy, 2001). We are testing candidate pathways that might mediate this signaling event, and looking at regulation of quartet-specific RNAs in response to organizer signaling.
The mesendodermal blast cell 4d: a hallmark of spiralian development.
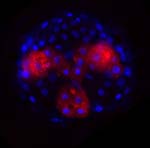
One of the most striking commonalities shared by the diverse animal phyla that have spiralian development is the 4d cell (Lambert, 2008). In spiralians, this cell divides bilaterally to produce a pair of large mother cells that give rise to a stereotyped series of daughter cells. This lineage gives rise to the intestine, and most of the mesodermal organs like the retractor muscle and the heart. We have recently found that the IoNanos mRNA is required for multiple tissues derived from 4d. This result is surprising, since Nanos is not known to play roles in somatic development outside of the insects. It suggests that a requirement for Nanos in the germline stem cells in the Drosophila ovary may extend to stem cell-like lineages in the soma as well. We are currently developing methods to determine the cleavage pattern of the 4d lineage, and the fates of particular cells in the lineage, as well as examining candidate genes from other systems.
Cited
Clement, A. C. (1962). Development of Ilyanassa following the removal of the D macromere at successive cleavage stages. Journal of Experimental Zoology 149, 193-216.
Halanych, K. M. (2004). The new view of animal phylogeny. Annual Review of Ecology Evolution and Systematics 35, 229-256.
Kingsley, E. P., Chan, X. Y., Duan, Y. and Lambert, J. D. (2007). Widespread RNA segregation in a spiralian embryo. Evolution & Development 9, 527-539.
Lambert, J. D. (2008). Mesoderm in spiralians: the organizer and the 4d cell. Journal of Experimental Zoology Part B-Molecular and Developmental Evolution 310B, 15-23.
Lambert, J. D. and Nagy, L. M. (2001). MAPK signaling by the D quadrant embryonic organizer of the mollusc Ilyanassa obsoleta. Development 128, 45-56.
Lambert, J. D. and Nagy, L. M. (2002). Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature 420, 682-6.
Sweet, H. C. (1998). Specification of first quartet micromeres in Ilyanassa involves inherited factors and position with respect to the inducing D macromere. Development 125, 4033-4044.