Cilia and cortical fibers in a vegetative Tetrahymena
visualized by selective anti-tubulin antibodies.
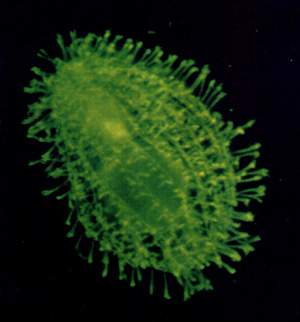
Two multi-gene families are under investigation in the Gorovsky lab: the histone genes and tubulin genes of the ciliated protozoan Tetrahymena thermophila. These studies have been greatly facilitated by the recent development of methods for mass transformation and gene replacement of the somatic macronucleus by the Gorovsky lab and by methods developed in collaboration with the laboratory of Peter Bruns (Cornell University) for transformation of the germline micronuclei. These methods now allow us to approach problems in Tetrahymena using many of the molecular genetic techniques that have been so successful in Saccharomyces cerevisiae. We are now engaged in applying these techniques to answer questions about aspects of these two multi-gene families which are highly conserved between Tetrahymena and multicellular eukaryotes but absent in yeast.
Our studies on histones emphasize their role in the regulation of gene expression and alteration of chromatin structure during mitosis. Macro- and micronuclei of Tetrahymena have a common origin during the sexual phase of the organism's life cycle. During vegetative growth they occupy the same cytoplasm but are extremely different in structure and function. Micronuclei are diploid, mitotically dividing, transcriptionally inert germinal nuclei. Macronuclei are endoreplicated, amitotically dividing, transcriptionally active somatic nuclei. This system presents a unique opportunity to study the mechanisms by which the same genetic information is differentiated and maintained in different functional states.
The total number of histone proteins and different histone genes in Tetrahymena is sufficiently small that a complete isolation and characterization of this gene family has been achieved. We are now restricting our analyses to histone subtypes that are found in almost all eukaryotes with the exception of S.cerevisiae. The H1-type histones of macro- and micronuclei are different, and there are two macronuclear-specific core histone variants. Recent studies have shown that either the gene encoding macronuclear H1 or the gene encoding the micronuclear linker histone (or both) can be knocked out without a detectable effect on growth. However, each knockout results in a nucleus-specific decondensation of chromatin, demonstrating for the first time that linker histones affect chromatin condensation in vivo. Current studies in this area center around determining the effects of H1 absence on transcription in vivo. Preliminary results indicate that the basal transcription level of a pol II gene is increased while its induced level of expression is unaffected. Interestingly, the induced level of expression of another gene is greatly reduced, suggesting that H1, like the core histones H3 and H4, can function in both gene activation and gene repression. Furthermore, we are initiating studies on the in vivo effects of phosphorylation of the linker histones at specific sites.
Using gene replacement techniques, we have shown that a conserved, quantitatively minor, macronuclear-specific H2A variant of the H2A.F/Z class (called hv1) is an essential gene while neither of the two major H2A genes is essential. We are now using these replacement techniques to analyze whether the essential function of this gene is due to the fact that it is constitutively expressed (i.e., is a basal or replacement variant) or due to its conserved primary sequence. Further studies on this variant center around testing the hypothesis that it functions to set up a transcriptionally competent chromatin state that enables the developmental removal of H1 from active genes (we and others have shown that active genes are deficient in H1). Presently, we are analyzing the function of specific modifications of this gene by acetylation and phosphorylation.
The final histone gene family we are analyzing contains the H3 genes. Tetrahymena has three H3 genes, two that encode the same major H3 protein and one that encodes a macronuclear-specific basal variant (hv2) that, like H3.3 of multicellular eukaryotes, is constitutively expressed. None of the three genes is essential. Surprisingly, when we knock out the gene encoding hv2, one of the two major H3 genes (which is normally expressed only in growing cells) is expressed constitutively. This result demonstrates that it is the constitutive expression, not the primary sequence of hv2 (which differs from the major H3 at 16 residues), that is essential. Based on the constitutive expression and the macronuclear specificity of hv2, we propose that all organisms need a constitutively expressed H3 gene to replace histone that turns over during transcription-a macronucleus-specific function. We are currently testing the hypothesis that in the absence of growth histone turnover occurs specifically in association with transcribed sequences. Other studies on the H3 gene family concern the role of micronuclear-specific phosphorylation of H3 in mitotic chromosome condensation.Intractyoplasmic and spindle microtubules in conjugating
Tetrahymena visualized by selective anti-tubulin antibodies.
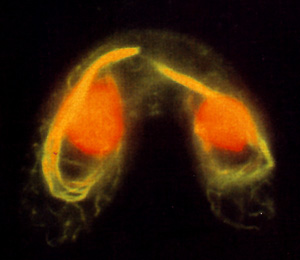 Studies with the tubulin gene system led us to clone and sequence the entire tubulin gene family of Tetrahymena, consisting of two beta-tubulin genes that encode an identical protein and one alpha-tubulin gene. Thus, the 17 distinct microtubule systems in Tetrahymena are produced from a single alpha-beta heterodimer, leading us to conclude that microtubule heterogeneity in this organism must be due to secondary modifications of tubulin or to microtubule associated proteins. We are now studying the effects of secondary modifications of tubulin using a method we developed that allows positive or negative selection for replacement of either the alpha- or beta-tubulin genes. This enables us to select directly for an otherwise wildtype tubulin gene containing a mutated site of secondary modification. Using this system we have shown that the highly conserved acetylation of Lys40 of alpha-tubulin is non-essential and that complete replacement with an alpha-tubulin that cannot be acetylated is without detectable phenotype. Currently, we are studying the function of the alpha-tubulin terminal tyrosine, the polyglutamylation of beta-tubulin, and the polyglycylation of both tubulins using a similar strategy. Our preliminary results suggest that both polyglutamylation and polyglycylation have essential functions which we are analyzing further.
Studies with the tubulin gene system led us to clone and sequence the entire tubulin gene family of Tetrahymena, consisting of two beta-tubulin genes that encode an identical protein and one alpha-tubulin gene. Thus, the 17 distinct microtubule systems in Tetrahymena are produced from a single alpha-beta heterodimer, leading us to conclude that microtubule heterogeneity in this organism must be due to secondary modifications of tubulin or to microtubule associated proteins. We are now studying the effects of secondary modifications of tubulin using a method we developed that allows positive or negative selection for replacement of either the alpha- or beta-tubulin genes. This enables us to select directly for an otherwise wildtype tubulin gene containing a mutated site of secondary modification. Using this system we have shown that the highly conserved acetylation of Lys40 of alpha-tubulin is non-essential and that complete replacement with an alpha-tubulin that cannot be acetylated is without detectable phenotype. Currently, we are studying the function of the alpha-tubulin terminal tyrosine, the polyglutamylation of beta-tubulin, and the polyglycylation of both tubulins using a similar strategy. Our preliminary results suggest that both polyglutamylation and polyglycylation have essential functions which we are analyzing further.
We are also studying the regulation of expression of the beta-tubulin genes. We have shown that each of the beta-tubulin genes is non-essential and that Tetrahymena responds to loss of one of its two tubulin genes by doubling the transcription from the remaining gene. We have shown that Tetrahymena has a unique signal transduction system in which any perturbation in the microtubule system, with either depolymerizing drugs or with taxol, results in the induction of tubulin transcription. Using tubulin mutations that alter the drug sensitivity of microtubules, we have shown that this system actually functions through the microtubules themselves. We have also shown that the initial four amino acids of beta-tubulin (MREI), which are conserved in almost all beta-tubulins and have been shown by Cleveland and colleagues to play an essential role in the post-transcriptional regulation of tubulin mRNA stability in mammalian cells, have no effect on tubulin mRNA amounts or stability in Tetrahymena. However, mutation of these residues causes dominant negative effects on cell growth, arguing that these sequences play an important role in the function of tubulin per se and that their role in regulation of mRNA stability is likely to have arisen later in evolution. We are presently trying to determine the mechanisms whereby perturbation of microtubules results in specific transcription induction of tubulin genes as well as the mechanisms underlying the ability of one beta-tubulin gene to be dosage compensated in the absence of the other one.
 Top of this page Back to Main Menu
Top of this page Back to Main Menu 
 Top of this page Back to Main Menu
Top of this page Back to Main Menu 

 Studies with the tubulin gene system led us to clone and sequence the entire tubulin gene family of Tetrahymena, consisting of two beta-tubulin genes that encode an identical protein and one alpha-tubulin gene. Thus, the 17 distinct microtubule systems in Tetrahymena are produced from a single alpha-beta heterodimer, leading us to conclude that microtubule heterogeneity in this organism must be due to secondary modifications of tubulin or to microtubule associated proteins. We are now studying the effects of secondary modifications of tubulin using a method we developed that allows positive or negative selection for replacement of either the alpha- or beta-tubulin genes. This enables us to select directly for an otherwise wildtype tubulin gene containing a mutated site of secondary modification. Using this system we have shown that the highly conserved acetylation of Lys40 of alpha-tubulin is non-essential and that complete replacement with an alpha-tubulin that cannot be acetylated is without detectable phenotype. Currently, we are studying the function of the alpha-tubulin terminal tyrosine, the polyglutamylation of beta-tubulin, and the polyglycylation of both tubulins using a similar strategy. Our preliminary results suggest that both polyglutamylation and polyglycylation have essential functions which we are analyzing further.
Studies with the tubulin gene system led us to clone and sequence the entire tubulin gene family of Tetrahymena, consisting of two beta-tubulin genes that encode an identical protein and one alpha-tubulin gene. Thus, the 17 distinct microtubule systems in Tetrahymena are produced from a single alpha-beta heterodimer, leading us to conclude that microtubule heterogeneity in this organism must be due to secondary modifications of tubulin or to microtubule associated proteins. We are now studying the effects of secondary modifications of tubulin using a method we developed that allows positive or negative selection for replacement of either the alpha- or beta-tubulin genes. This enables us to select directly for an otherwise wildtype tubulin gene containing a mutated site of secondary modification. Using this system we have shown that the highly conserved acetylation of Lys40 of alpha-tubulin is non-essential and that complete replacement with an alpha-tubulin that cannot be acetylated is without detectable phenotype. Currently, we are studying the function of the alpha-tubulin terminal tyrosine, the polyglutamylation of beta-tubulin, and the polyglycylation of both tubulins using a similar strategy. Our preliminary results suggest that both polyglutamylation and polyglycylation have essential functions which we are analyzing further. Top of this page Back to Main Menu
Top of this page Back to Main Menu 